
Ciprofloxacin Triggers Lasting Resistance Across Gut Bacteria
Stanford University researchers have found that even short-term use of ciprofloxacin can drive persistent antibiotic resistance in the human gut microbiome. Resistance emerged independently across multiple bacterial species and remained detectable for over 10 weeks after treatment ended.
A New Approach to Studying Antimicrobial Resistance
While antimicrobial resistance (AMR) continues to threaten global health, much previous research has relied on in vitro models or animals, which cannot fully capture the complexity of human microbial environments. To address this gap, Stanford scientists designed a longitudinal metagenomic study titled “Brief antibiotic use drives human gut bacteria towards low-cost resistance,” published in Nature.
Study Design: Tracking Resistance Over 20 Weeks
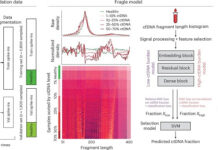
Researchers enrolled 60 healthy adults who received ciprofloxacin, 500 mg twice daily, for five days. Over the following 20 weeks, participants collected 16 stool samples each, resulting in 960 samples for analysis. The team performed shotgun metagenomic sequencing, generating an average of 18.8 million reads per sample. They also developed a computational tool, PolyPanner, to identify true polymorphic sites over time.
Key Findings: Rapid and Widespread Evolution
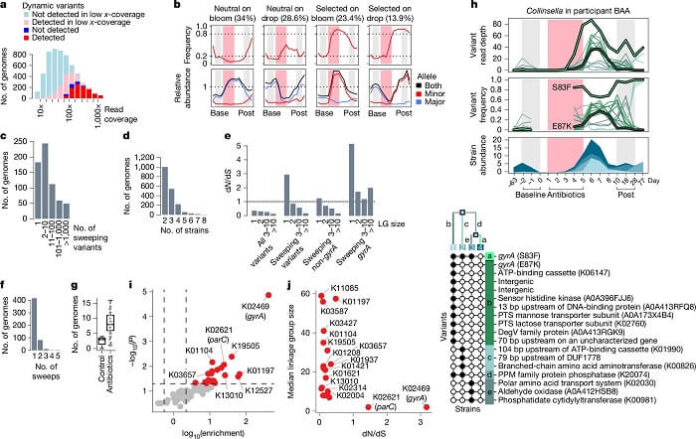
As reported by medicalxpress, the study reconstructed 5,665 bacterial genomes from the stool samples and identified 2.3 million genetic variants. Out of these, 513 bacterial populations showed selective sweeps—clear indicators of adaptive evolution. Researchers found a high concentration of mutations in the gyrA gene, a known marker for fluoroquinolone resistance.
Specifically, 63 populations across 34 participants developed gyrA mutations, typically arising independently within each individual. Nearly 10% of previously susceptible bacterial populations evolved resistance during the study period.
Resistance Persists Without Fitness Cost
Once these gyrA mutations appeared, they persisted for more than 10 weeks and are predicted to remain for up to a year. Importantly, these resistance mutations did not impose a fitness cost, meaning resistant bacteria continued to dominate even after antibiotic treatment ended. Targeted sequencing revealed no evidence of resistance reversion, suggesting long-lasting genetic changes.
Although most mutations occurred in gyrA, researchers also identified resistance-associated mutations in other genes, albeit less frequently.
Factors Influencing Resistance Development
Resistance was more likely to develop in bacterial populations that were abundant before treatment and suffered sharp declines during antibiotic exposure. This pattern highlights the role of population dynamics in predicting evolutionary changes.
Moreover, gut bacteria evolved resistance without needing prior infections, indicating that commensal microbes could act as reservoirs for resistance traits. These traits could later transfer to pathogenic bacteria through horizontal gene transfer.
Future Directions: Toward Predictive Resistance Models
Given that resistance evolution correlated with starting population sizes, researchers see potential for predictive models. By monitoring microbial composition and abundance before and during antibiotic treatment, clinicians could anticipate resistance risks and tailor therapies accordingly.
However, the study emphasizes the need for further experiments using different population mixes and antibiotic types to refine predictive strategies.
Conclusion: Rethinking Antibiotic Stewardship
This research highlights that even brief antibiotic courses can cause long-lasting changes to the human gut microbiome. It underscores the urgent need to monitor microbial populations closely during treatment, optimize antibiotic usage, and implement precision-guided stewardship practices to combat the growing threat of AMR.